Precision Neural Engineering Lab
MR.Flow
MRI processing pipeline orchestration systemMR.Flow is a workflow orchestration system for MRI data processing—managing multi-step analysis pipelines across subjects. Built to handle sequential processing stages (quality control → preprocessing → statistical analysis) with worker coordination, task queuing, and failure recovery.
The system orchestrates complex neuroimaging workflows involving multiple processing tools (fMRIPrep, FSL, custom QC), tracks job state across distributed workers, and manages dependencies between processing stages. Designed for research environments where hundreds of subjects need consistent, reproducible processing pipelines.
A pipeline orchestration platform for neuroimaging research—task scheduling, worker coordination, dependency management, and failure recovery for multi-stage MRI analysis workflows.
MR.Flow — Visual pipeline orchestration for neuroimaging research
Pipeline Architecture
Core Components
What Problem It Solves
Most MRI tools assume:
- You already understand every container, flag, path, and failure mode
- You'll debug everything manually
- You'll glue ten tools together yourself
- Installing tools (days of work) and BIDS formatting (terminal fluency required) are your problem
For a study with 150+ datasets, this meant weeks of manual work, constant babysitting of jobs, and high risk of human error in data organization.
MR.Flow replaces that entire failure-prone layer with a visual command center.
What Makes MR.Flow Different
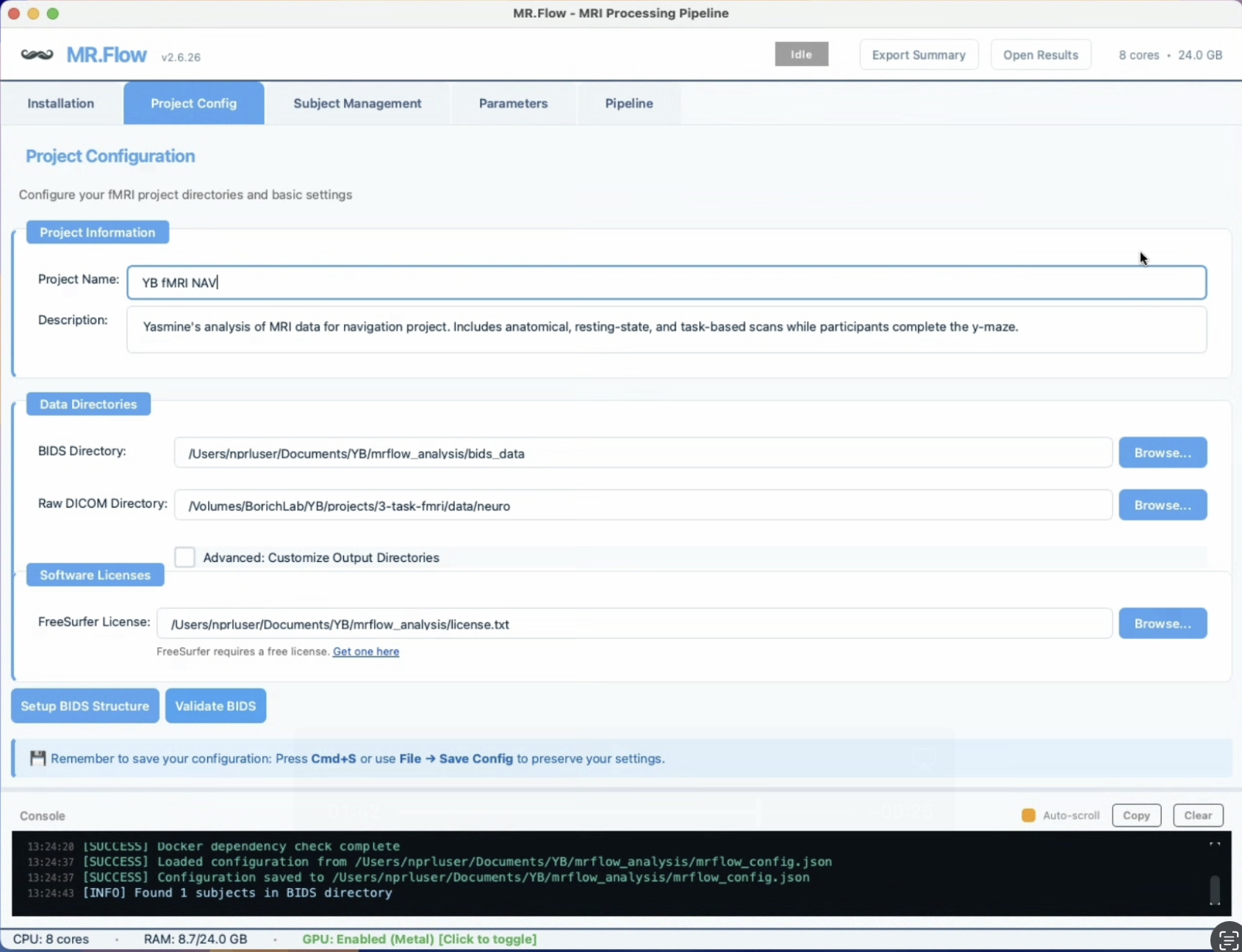
One-Click Orchestration of fMRIPrep, MRIQC, FreeSurfer, XCP-D, FSL
- One-click orchestration of fMRIPrep, MRIQC, FreeSurfer, XCP-D, FSL
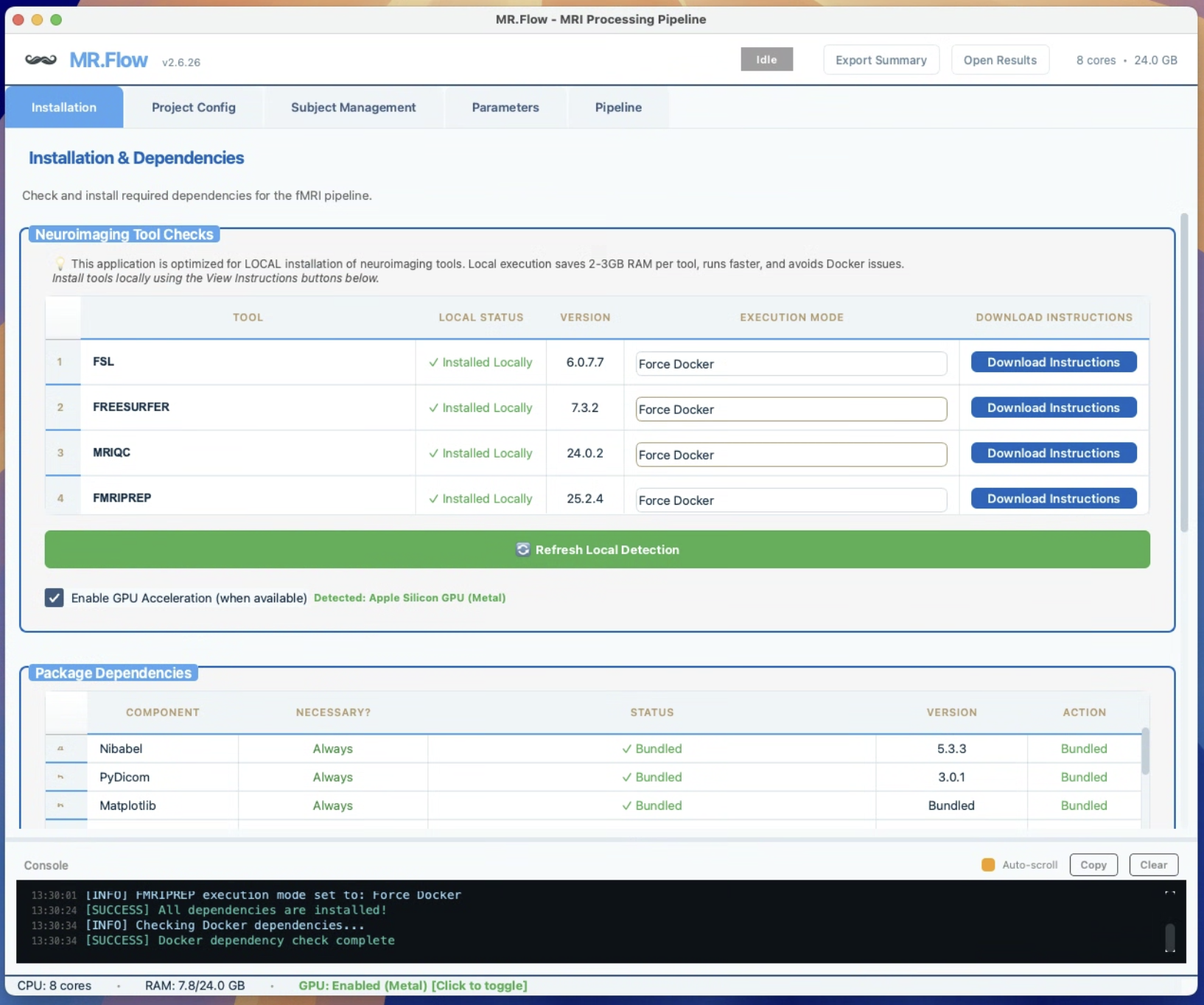
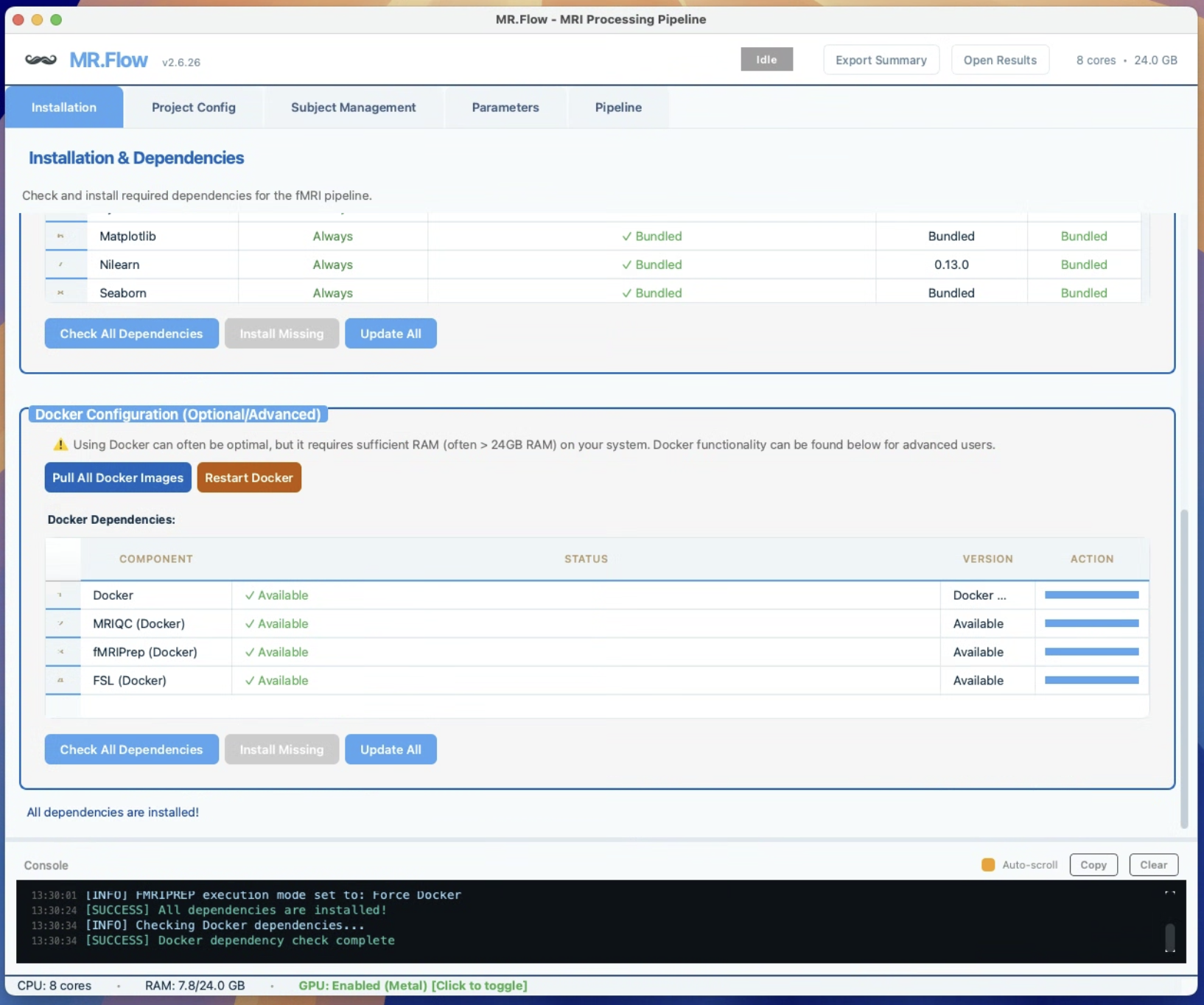
- Automatic Docker detection & launch — starts Docker for you on macOS, detects installed images, handles platform mismatches (Apple Silicon vs AMD64)
- Staged pipeline with dependency tracking — interrupted jobs resume from where they stopped, not from the beginning
BIDS Validation with Auto-Healing for Missing Metadata
- BIDS validation & self-healing — detects missing dataset_description.json, auto-generates missing sidecar JSONs, flags short runs and missing metadata
- Data integrity checks before compute — catch problems before they corrupt results
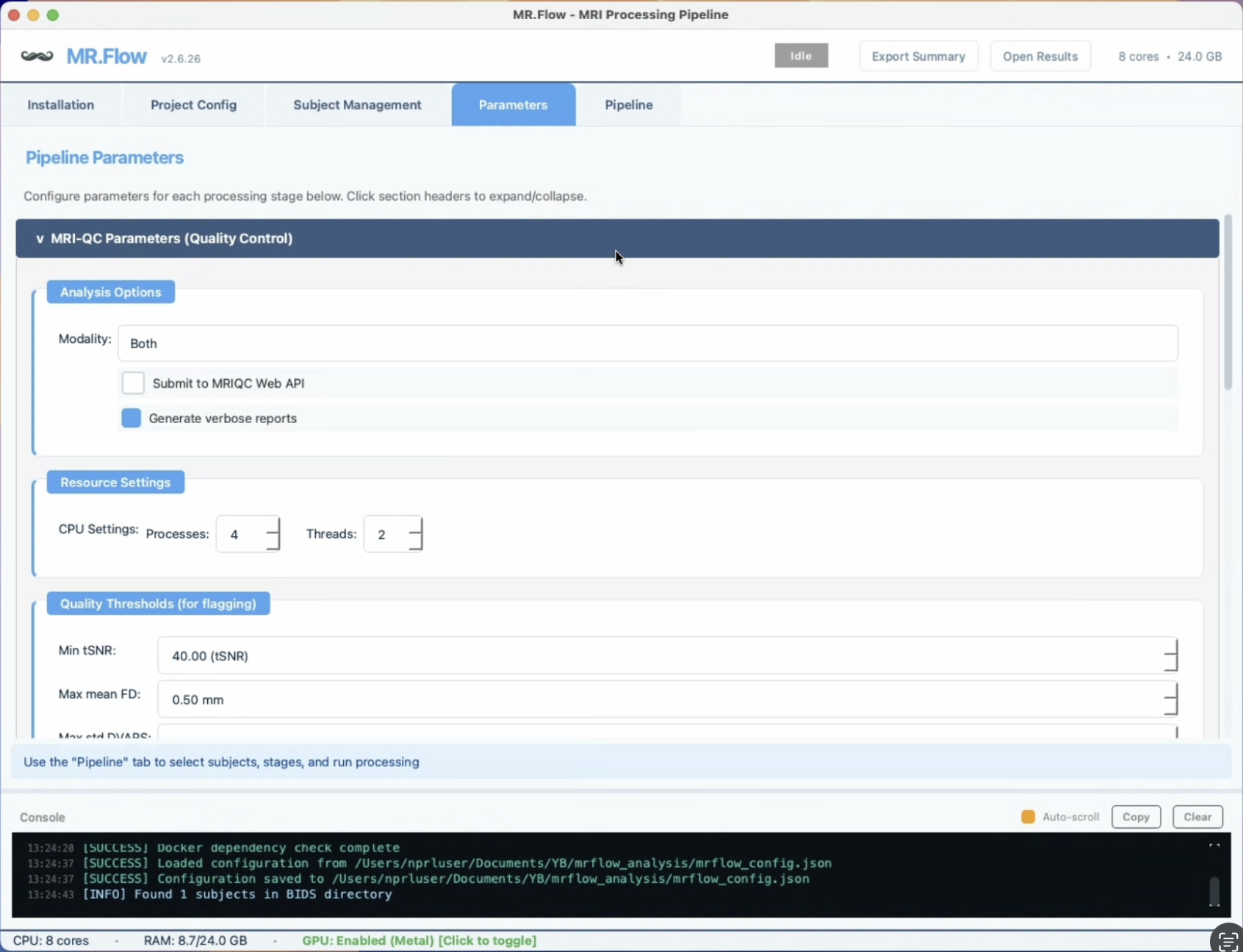
Integrated MRIQC with Automated Outlier Flagging
- Integrated MRIQC — tSNR, FD, DVARS, ghosting checks
- Automatic outlier flagging with visual QC tables and export
- No more QC blindness — problems surface immediately, not after weeks of processing
Hardware-Optimized Parallel Execution
- System-aware resource scaling — detects CPU cores, RAM, GPU availability
- Auto-suggests parallelization settings based on your hardware
- Intelligent work queue distribution across available resources
- Persistent project state — every project saved as structured JSON, reloadable across sessions
- Live pipeline console — all subprocess output captured, errors/warnings/logs structured in one place
- No more lost terminal history — deterministic pipelines instead of shell-script entropy
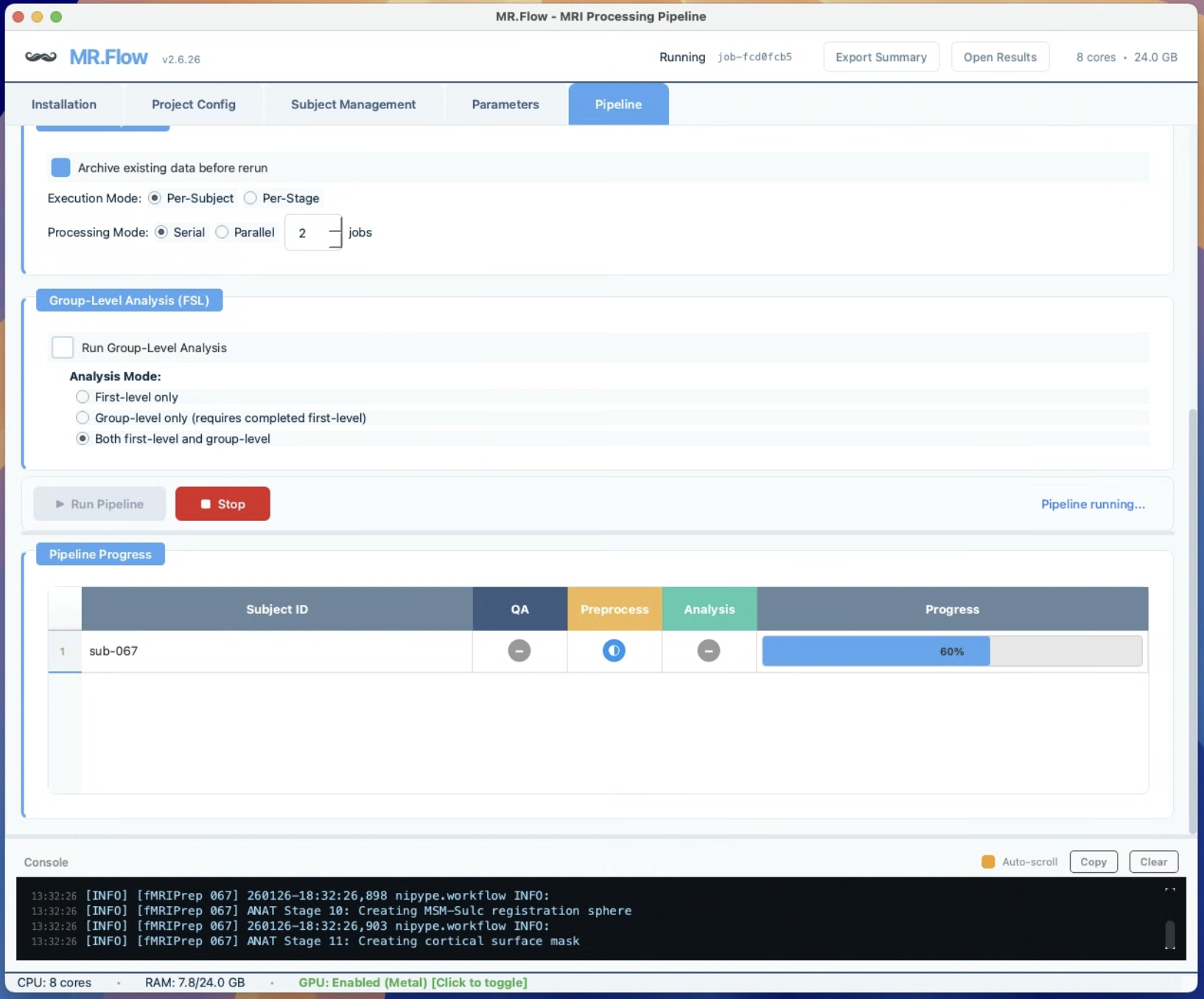
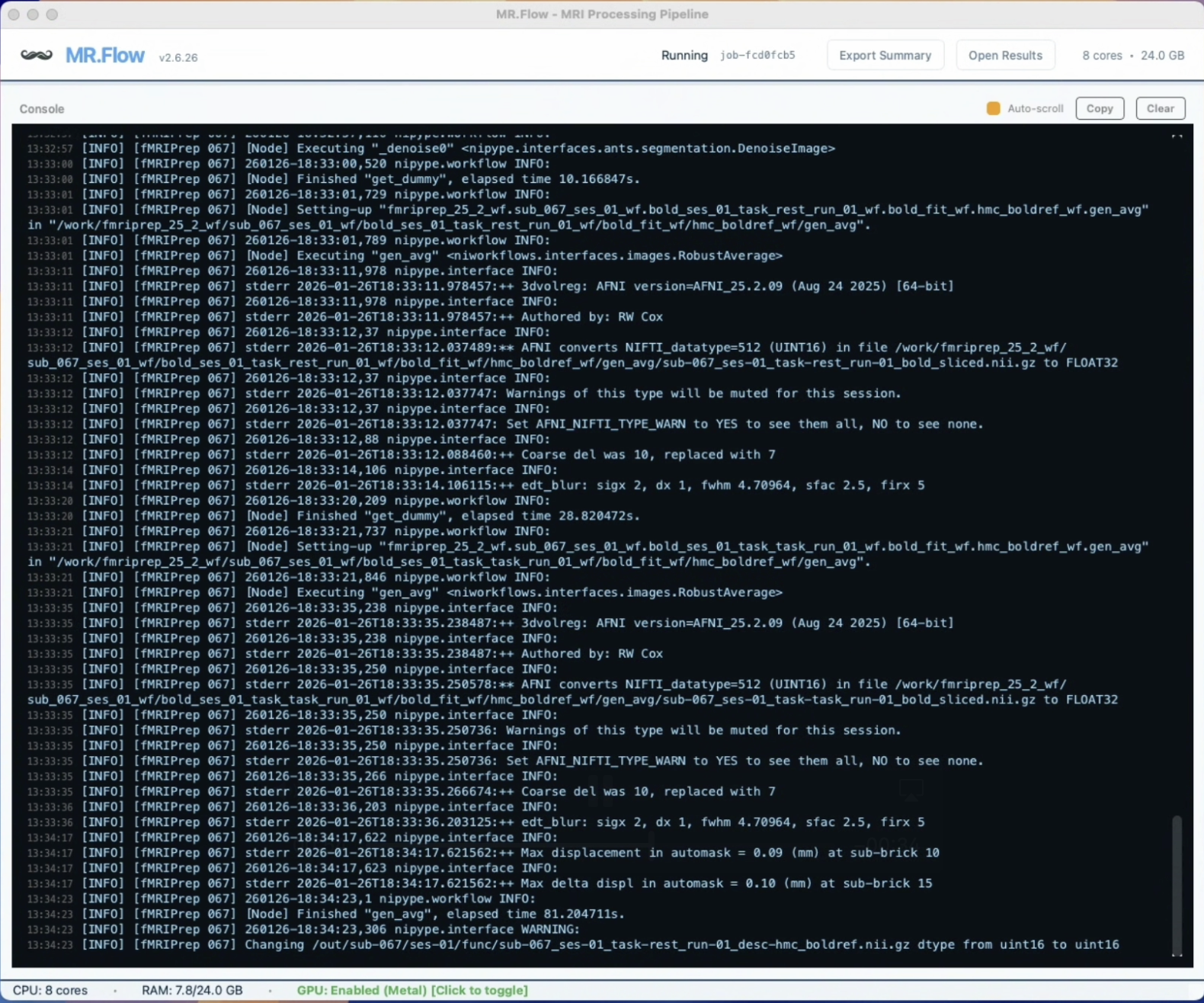
Handles Clinical Data with Missing Files and Lesions
- Handles imperfect datasets — built for real-world data with missing files, inconsistent naming, and incomplete metadata
- Clinical population support — processes atypical images (e.g., stroke patients with lesions) that often break traditional pipelines
- Multi-modal scan detection — automatic identification of anatomical, functional, resting state, and diffusion sequences
- Graceful degradation — continues processing what's available rather than failing on the first anomaly
Designed for How Scientists Actually Work
MR.Flow is not just a launcher—it is a thinking environment for neuroimaging workflows:
- You configure experiments visually
- You validate assumptions before you compute
- You catch data integrity problems before they corrupt results
- You get deterministic pipelines instead of shell-script entropy
Why This Exists
MR.Flow grew out of real clinical research failures—missing metadata, broken Docker images, silent QC failures, and weeks lost to debugging shell scripts. It is built to make MRI pipelines survivable for humans.
Platform lesson: Scientists don't want to understand Docker, BIDS validation, or container orchestration—they want their research to work. The same way developers don't want to understand infrastructure complexity—they want their apps deployed. MR.Flow is a control plane for neuroimaging.
Impact
- Reduced preprocessing time from weeks of manual work → days automated
- Eliminated human error in data organization and file naming
- Enabled reproducible science across the lab
- Deployed in production for active neuroimaging research across multiple studies
- Used by researchers with varying technical backgrounds
Who It's For
- Cognitive neuroscience labs
- Clinical imaging studies
- Graduate students drowning in preprocessing
- PIs who want reproducibility without DevOps
- Anyone tired of fragile shell scripts holding up million-dollar scanners
How does this connect? MR.Flow exemplifies how accidental architecture prevents processing—and how redesigning that architecture enables exploration. See how this fits the larger pattern →