Precision Neural Engineering Lab
Epoche
ML workbench for neurophysiology researchEpoche is an ML workbench for neurophysiology—signal processing, feature extraction, hyperparameter exploration, ensemble optimization, cross-model interpretability, and publication-ready export, all through a visual interface.
Designed for closed-loop neuroscience paradigms where trained models must be deployed to real-time systems. Models developed in Epoche are deployed to my C++ real-time execution backend with ~40ms classification latency, integrating multiple data streams via Lab Streaming Layer (LSL) and controlling experimental paradigms through analog/digital triggers for EEG, EMG, robotics, and neurostimulation.
A complete ML workbench for neurophysiology—from raw signals to trained models, interpretability narratives, and publication-ready reports. Everything configurable through a GUI, reproducible by default, and deployable to real-time closed-loop systems.
Epoche — Visual EEG-to-model classification pipeline
Platform Architecture
ML Pipeline Flow
Engineering Challenge
The problem: neuroscience experiments need ML models that can classify brain signals in real-time, but typical ML workflows focus on offline analysis, offer no interpretability, and leave researchers writing throwaway scripts.
- Latency requirements — classifications must complete in sub-milliseconds for closed-loop systems
- Real-time feature extraction — compute signal features from streaming data without batching delays
- Model selection at scale — comparing 18 model types × hyperparameter spaces × ensemble combinations is intractable without tooling
- Interpretability gap — accuracy numbers alone don't explain why a model works or generate mechanistic hypotheses
- Model deployment — trained models must be serializable and loadable in real-time C++ systems
- Reproducibility — same preprocessing, features, and model configuration in training and inference
The solution: a visual workbench that handles the full lifecycle—exploration, optimization, interpretation, and export—with identical feature pipelines between training and deployment.
What Problem It Solves
Most EEG classification workflows assume:
- You'll write throwaway scripts for each experiment
- You'll reinvent preprocessing, feature extraction, and evaluation every time
- Reproducibility is your problem—buried in ad-hoc code
- Models stay in Python—real-time deployment is someone else's problem
This creates fragile pipelines, inconsistent results, and a gap between model development and production deployment.
Epoche bridges offline model development with real-time deployment—visual configuration, unified hyperparameter/ensemble exploration, cross-model interpretability, and serialization for production execution.
What Makes Epoche Different
Visual Configuration — GUI-Driven, No Code Required
- GUI-driven pipeline — configure the entire workflow visually
- No code required — researchers can iterate without engineering support
- Reproducible by design — every run captures its exact configuration
Signal Processing — Multi-Format Ingestion with Quality Metrics
- Multi-format ingestion — EDF, BDF, CSV supported out of the box
- Preprocessing pipeline — filtering, resampling, epoch extraction
- Signal quality metrics — automated artifact detection before you waste compute
9 Feature Categories with 100+ Extraction Methods
- Time-domain — basic statistics, Hjorth parameters (activity, mobility, complexity)
- Frequency-domain — FFT, power spectral density, spectral entropy
- Time-frequency — wavelet decomposition, wavelet energy/entropy
- Complexity measures — sample entropy, permutation entropy, fractal dimension
- Nonlinear dynamics — DFA, Hurst exponent, Lyapunov, correlation dimension
- Spatial features — Common Spatial Patterns (CSP), Riemannian tangent space, coherence/PLV
- Dynamic feature registry — categories enable/disable based on installed dependencies, with NumPy fallbacks for optional packages
18 ML Models from Logistic Regression to Deep Ensembles
- 18 model types — classification and regression through a unified API (scikit-learn compatible)
- Automated cross-validation — configurable k-fold strategies without boilerplate
- Comparative visualization — evaluate model performance side by side
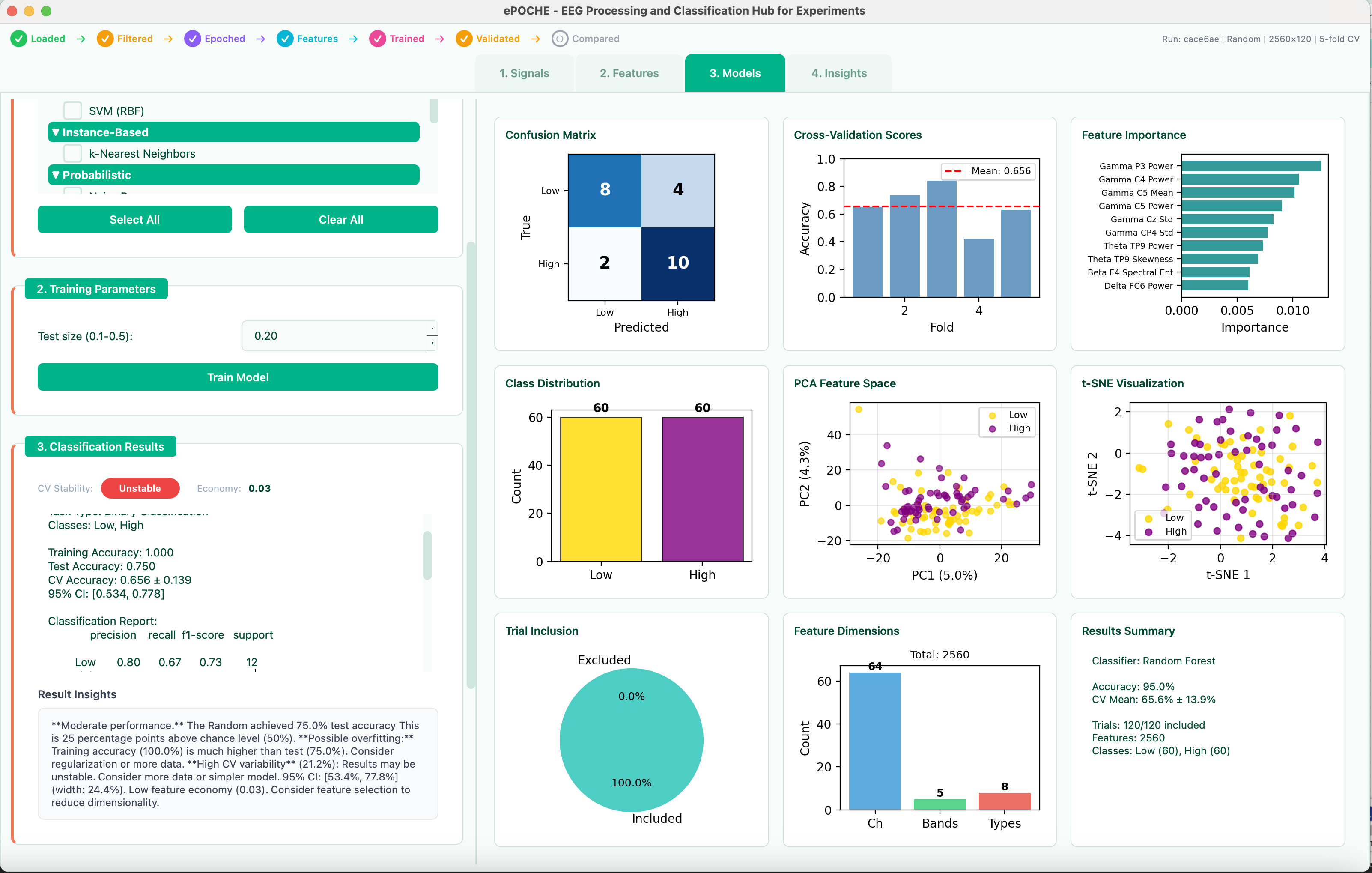
Unified Grid Search Across All Models + Ensembles
- Unified grid search — select any combination of models, set each hyperparameter to fixed or explore (range/list), and search the full space in one run
- Ensemble search — automatically tests all C(n,k) combinations of top models across voting (hard/soft/weighted) and stacking strategies with configurable meta-learners
- Visual analysis — parameter sensitivity heatmaps, Pareto frontier (accuracy vs. complexity vs. speed), and ranked ensemble tables
- Persistent results — explorations save to SQLite with full result blobs for reload and comparison
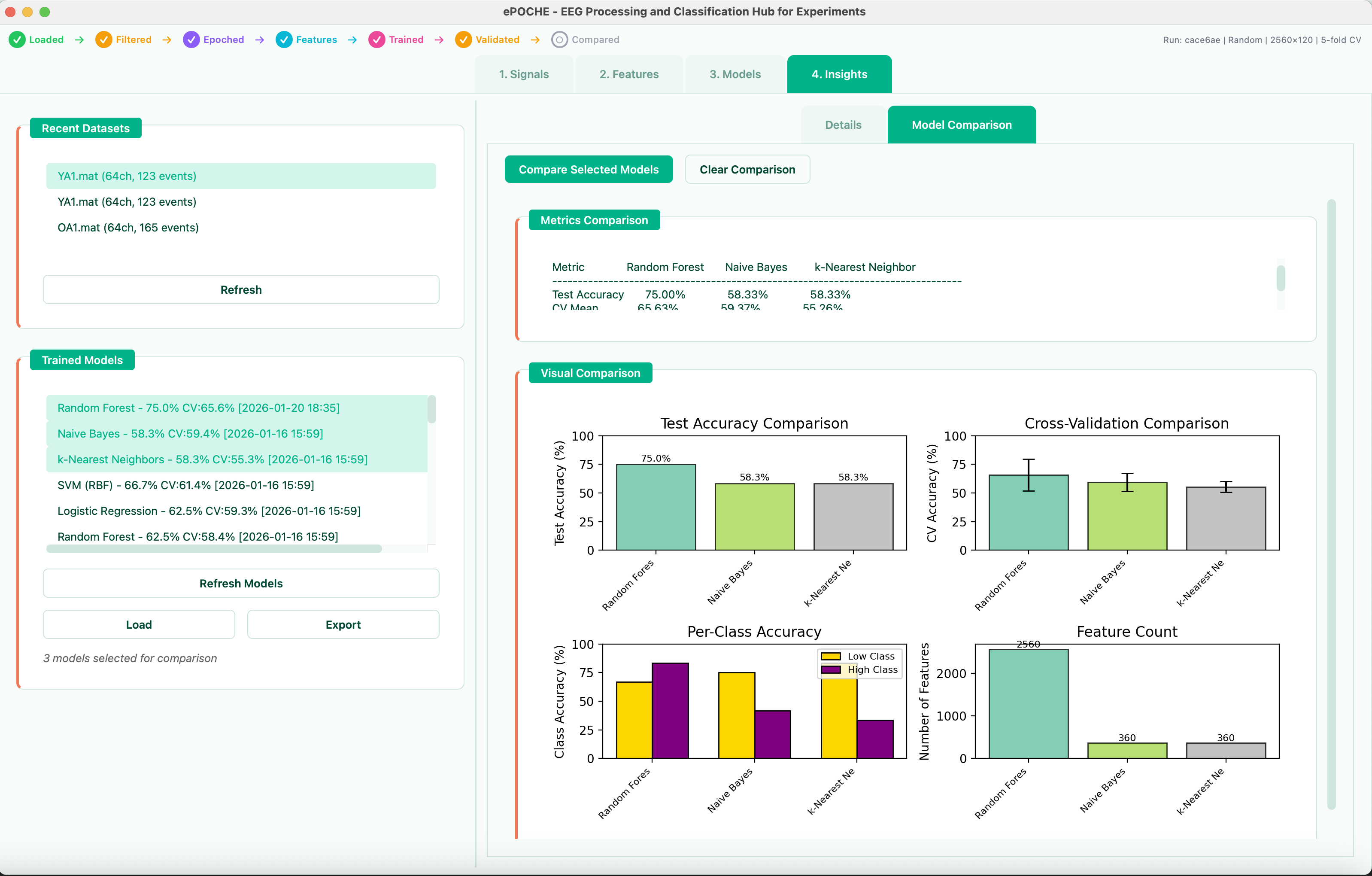
7 Interpretability Methods with Cross-Model Consensus
- 7 analysis methods — SHAP (global + instance), LIME, permutation importance, decision rules, unified importance profiles, and cross-model comparison
- Cross-model consensus — Spearman rank correlations, Friedman test, per-feature coefficient of variation across models
- Hypothesis generation — auto-detects divergence patterns (tree-vs-linear, single-model-high, band-specific) and generates testable hypotheses with EEG context
- EEG-specific visualizations — topographic maps (10-20 system), frequency band importance aggregation, spatial comparison across datasets
- Cross-dataset comparison — load two saved interpretations, compare importance patterns with Pearson/Spearman correlations, identify stable vs. dataset-specific features
Publication-Ready Reports + Real-Time C++ Deployment
- Publication-ready reports — export comparison results as PDF (multi-page via matplotlib), self-contained HTML (base64-embedded figures), or LaTeX source with figure files
- Model serialization — pickle-serialized models with preprocessing pipelines for deployment
- Real-time backend integration — exported models deploy to C++ execution with sub-millisecond latency
- Reproducibility logs — every parameter, feature set, and model configuration captured for exact replication
Designed for How Researchers Actually Work
Epoche is not just a script collection—it is a thinking environment for biosignal classification:
- You explore data before committing to a model
- You iterate on features without rewriting pipelines
- You search hyperparameter spaces and ensemble combinations in one unified exploration
- You interpret results across models—consensus, divergence, and mechanistic hypotheses
- You export publication-ready reports that reviewers can reproduce
Why This Exists
Epoche grew out of watching researchers rebuild the same infrastructure for every EEG study—loading data, extracting features, training models, generating figures. The exact pipeline was always buried in scripts that worked once and broke forever.
The abstraction layer means the same tool works for EMG, ECG, GSR—any biosignal classification. Swap data sources and model types without touching underlying code.
Platform lesson: Researchers don't want to rebuild the same ML pipeline infrastructure for every study—they want to iterate on classification approaches, understand why models disagree, and generate hypotheses. Epoche is a control plane for biosignal classification: exploration, interpretation, and deployment through one interface.
Impact
- Reduced pipeline setup from days to hours
- Enabled systematic comparison of classification approaches across models, hyperparameters, and ensembles
- Interpretability as a first-class feature — researchers get mechanistic hypotheses, not just accuracy numbers
- Made results reproducible by default with publication-ready export
- Accessible to researchers without ML engineering background
Who It's For
- Cognitive neuroscience researchers
- BCI developers prototyping classification
- Graduate students learning ML for biosignals
- Anyone who needs reproducible EEG/EMG/ECG analysis
How does this connect? Epoche treats model architecture as a variable to understand what patterns are real vs architectural artifacts. See how this connects to the broader question →